Infertility
Echographic call signs and ACPA (chromosome analysis on DNA chip)
*Article written in 2017*
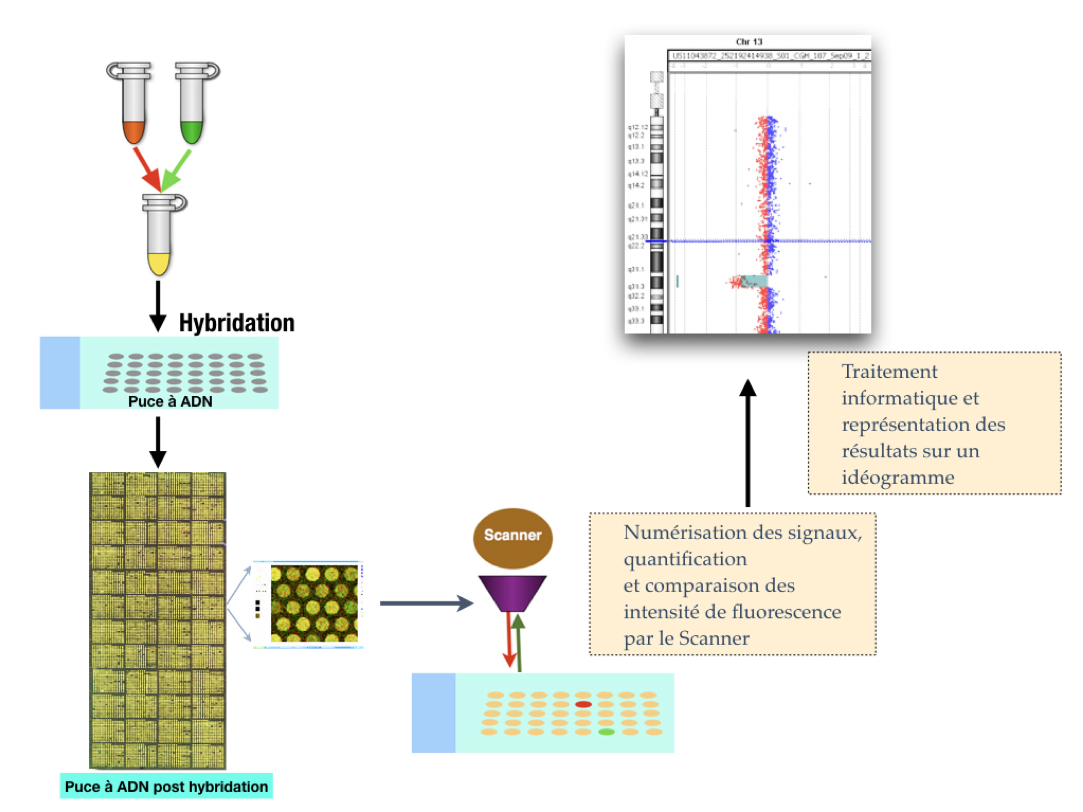
In the late 1990s, sequencing of the human genome produced sequenced DNA fragments that were cloned in bacteria or yeast. In the late 1990s, Pinkel et al. and Solinas-Toldo et al. had the idea to deposit these DNA sequences with known genome positions on glass slides(1,2). This is how the first DNA chips or "microarray" appeared and the technique of CGH array developed. Currently, oligonucleotides (or "probes") of about 60 bases are fixed on the blades. Depending on the number of probes chosen, the resolution of the chip may vary from about 1 Mb (megabyte) to 1 kb (kilobase).
CGH array principle
Its principle is to co-hybridize the same amount of DNA from a patient and control, each marked by a different fluorochrome, to a blade on which probes are attached. After a hybridization step, the signals generated by the two fluorochromes are digitized and a ratio of their respective intensity (reflecting the ratio of the patient’s DNA quantity to that of the control) is established at each locus. A graphical representation of the anomaly is obtained through software.
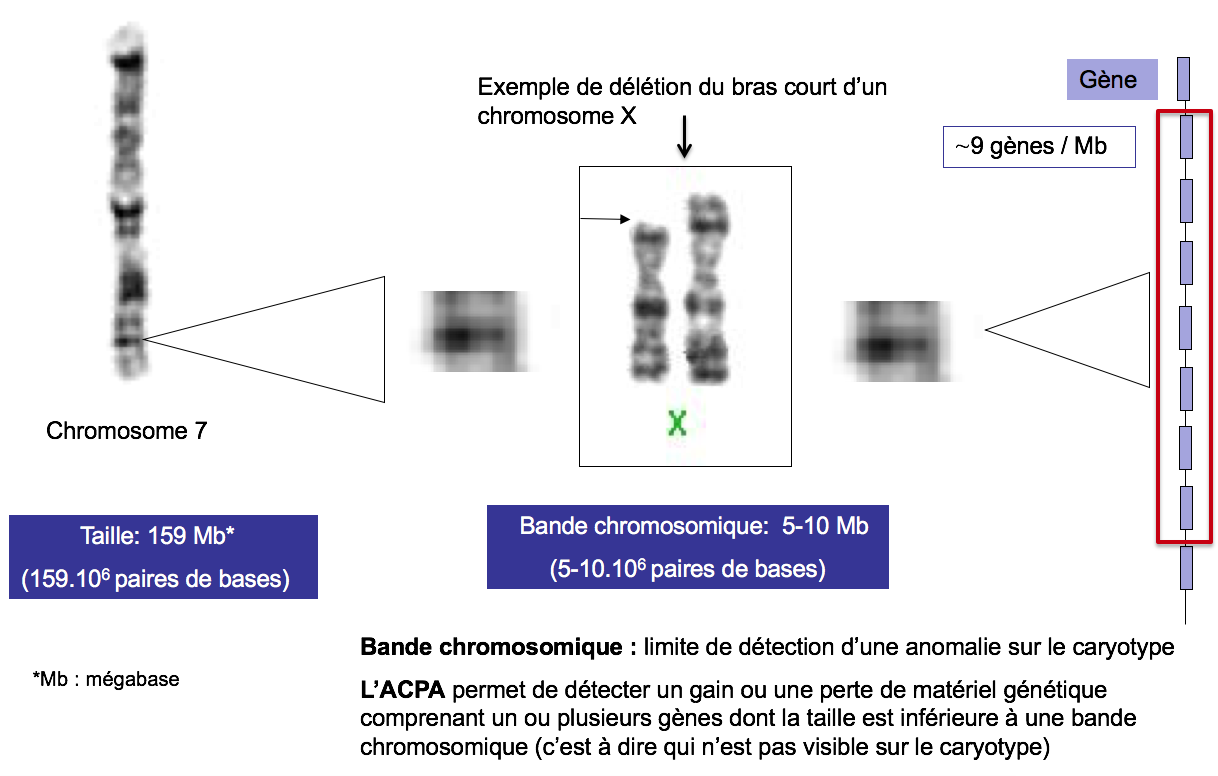
With this new technology, it was possible to detect genomic imbalances that are not visible on the karyotype, i.e., smaller than 5-10 Mb (which corresponds to the size of a chromosome band) (Figure 1).
Subsequently, other types of chips, known as SNP chips or "SNP array" (single nucleotide polymorphism), were developed. These are chips that use short and very specific oligonucleotides that can distinguish sequences that differ from only one nucleotide. On average, one SNP is met every 100 to 1,000 nucleotides. Their number is about 5.106 in the human genome. SNP chips offer the advantage of detecting, in addition to genomic imbalances, situations of single-parent disomy (two chromosomes from the same parent). In France, the term DNA chip chromosome analysis (ACPA) was chosen by the French network "AChro-Puce" to refer to both the techniques of CGH (comparative genomic hybridization) array or comparative genomic hybridization chip as well as the techniques of SNP array (http://www.renapa.univ-montp1.fr/).
Figure 1. Example of the deletion of the short arm of a X Chromosome 7
Size: 159 Mb* (159.106 base pairs)
Chromosome band: 5-10 Mb (5-10.106 base pairs)
Chromosome band: limit of detection of an anomaly on the karyotype
ACPA allows to detect a gain or loss of genetic material comprising one or more genes whose size is less than a chromosome band (i.e. not visible on the karyotype).
ACPA Medical Applications
The development of the ACPA technique (or chromosome analysis on a DNA chip) has therefore led to a paradigm shift in medical cytogenetics. The first applications of ACPA concerned the cytogenetics of solid tumors due to the complexity of abnormalities detected on the karyotype. Constitutional pathology has been the second field of medical application of this technique with the exploration of patients with isolated or syndromic intellectual deficits. Various studies have shown that ACPA can detect approximately 12% of cryptic genomic imbalances (i.e., not visible on karyotype) in these patients(3). For this reason, this technique has become the first-line examination for genomic analysis of patients with intellectual deficiency and/or birth defects. However, the clinical impact of an Anomaly identified by ACPA may be difficult to determine, constituting a major impediment to the application of this technique in prenatal. The technical means pre-empted our understanding of the medical implications of the data generated.
CNVs (copy number variants): classification
The genomic imbalances identified by ACPA are called CNVs (copy number variants) and correspond to quantitative variations of the genome compared to a reference genome*.
It is important to note that a CNV is only a loss or gain of chromosome material without being able to predict its pathogenicity. In fact, a CNV can be mild, i.e. without phenotypic consequences for the individual carrying it. Currently, it is estimated that approximately 16% of the genome would correspond to non-deleterious variations(4). However, it has been shown that some of these so-called benign CNVs have a role in the immunity or occurrence of common pathology such as systemic lupus(5,6).
In addition to these benign CNVs, there are pathogenic CNVs because they cause neuro-developmental disorders. Many syndromes, such as the 17q21.31 (Koolen-de-Vries syndrome), could be described as a result of the generalization of postnatal ACPA(7). It should be noted that the majority of pathogenic CNVs are not recurrent and are scattered across the chromosomes. Thus, a penomic and non-targeted approach is needed to detect them.
In addition to these clear situations, there are CNVs that pose genetic counseling problems. On the one hand, these are CNVs for predisposition to the occurrence of neuro-developmental pathologies. For example, 16p11.2 deletion or 1q21.1 duplication associated with autistic spectrum disorders(8,9). Case-control studies have shown that these CNVs are susceptibility factors to these conditions because they are more frequently detected in patients than in controls. However, at present, this dichotomy between healthy individuals and controls is not considered to reflect reality, as there is in fact a very broad phenotypic spectrum in individuals with this type of CNV. In fact, a minima cognitive impairment was detected retrospectively in carrier parents when initially considered healthy(10,11). To date, the genetic, epigenetic, and/or environmental factors leading to the expression of disease are not known. Genetic counseling is therefore very difficult in antenatal because it is not possible to predict the phenotype of a fetus carrying this type of CNV.
In addition, the second category of CNV that raises genetic counseling problems corresponds to SUVs (variation of certain significance, or variant of uncertain meaning). These are "private" and often inherited CNVs whose clinical consequences for the developing fetus are not established. This creates great anxiety when the information is passed on to the parents and can lead to an unjustified request for a medical interruption of pregnancy.
Finally, exceptionally, a predisposing CNV at the onset of a late revelation pathology can also be identified(12). The application of this new technology in prenatal care should therefore lead to a consideration of the ethical problems it raises.
Due to the difficulty of interpreting some CNVs (SUVs) or the possible detection of a CNV that has a medical impact that is not related to the initial indication ("Incidental discovery CNVs"), international recommendations call for a pre-test consultation to be offered to the patient so that accurate and clear information can be provided by trained health personnel (doctor, doctor, midwife, genetic counselor...)(13,14). The gathering of specific consent for this analysis is essential. Finally, when a CNV is notified on the report, it must be explained to the patient during a post-test consultation.
Benefits and limitations of prenatal ACPA
In recent years, the feasibility and benefits of applying ACPA in prenatal have been widely demonstrated(15). The main advantages of this prenatal technique are:
- the possibility of detecting cryptic pathogenic CNVs;
- faster detection than the non-cryptic CNV karyotype, as a result can be achieved in less than a week by the fact that the technique is carried out on uncultivated cells (from the collection of amniotic fluid, choral villi or fetal blood);
- the possibility of analyzing cells that are not capable of dividing. An ACPA can be performed from fetal tissues (skin, lung, thymus..) after a fetal death in utero for example;
- a fine characterization of the genomic imbalance identified (size of the CNV and content of genes). The molecular characterization of a chromosome reshuffle allows a more accurate phenotype - genotype correlation to be established and thus provides more adequate genetic advice.
While CAPA is a high-performance technology, it is important to note that it has limitations. Since the principle of the technique is to compare the amount of DNA of a patient with a witness, balanced reshuffles (without loss or gain of chromosome material) are not highlighted. Moreover, cell-by-cell analysis is not possible, unlike karyotype or FISH (fluorescence in situ hybridization) and, as a result, low mosaics (< 10-20%) are not detected. Finally, triploidia can only be detected by SNP chips and not by CGH array.
Another limitation to be considered is that the ACPA provides quantitative information without specifying the type of anomaly identified (e.g., translocation or insertion derivative). The identification of a CNV should therefore be supplemented by a FISH study. In addition, a FISH study in parents is required to determine the character of the anomaly de novo or inherited, which in some situations will assist in the classification of the CNV (pathogen or non-pathogen).
Indications of prenatal ACPA
A prenatal chromosome diagnosis is proposed where there is a significant increase in the risk of unbalanced chromosomal abnormality in the fetus that could lead to sufficiently serious and/or lethal developmental disorders to propose a medical interruption of pregnancy. Various studies have shown that the rate of cryptic CNV pathogens detected by ACPA is about 9% in polymalformed fetuses. In the case of isolated malformation, this rate is lower at around 6%(16). For example, the realization of a penomics ACPA in fetuses with morphological abnormality(s) is a diagnostic strategy that has been adopted in many countries, notably in France (ACLF Recommendations http://www.eaclf.org/docs/ ACPA/GBP_ACPA-v10.pdf). According to Wit et al. metaanalysis, cardiac, central nervous system, and musculoskeletal system malformations are the most important sources of pathogenic chromosome abnormalities(16).
ACPA in fetuses with heart disease
Heart disease is one of the most common birth defects at birth. Their prevalence is estimated to be between 4 and 8 per 1,000 live births. In antenatal, the incidence of chromosome abnormalities in fetuses with heart disease is high, about 25%(17). These are mainly cases of trisomy 21, trisomy 18, trisomy 13, monosomy X and microdeletion 22q11.21. A recent meta-analysis of Jansen et al. including 1131 fetuses with cardiac disease revealed that ACPA detected approximately 7% of abnormalities (after excluding aneuploidy and microdeletion cases 22q11.21)(18). This rate is higher in syndromic forms (9.3%) than in isolated forms (3.4%). In addition, chromosome abnormality is more frequently found in the case of syndromic interventricular communication and left ventricular ejection pathway abnormalities. It should be noted that pathogenic CNVs have also been identified in fetuses with large vessel transposing or heterotaxis. Currently, it is therefore recommended to perform ACPA regardless of the type of heart disease(18-20).
ACPA in fetuses with a central nervous system defect
Central nervous system abnormalities are often severe and are a frequent cause of medical termination of pregnancy(21). They concern 0.14-0.16% of live births and 2-6% of stillborn children. The rate of pathogenic CNVs in fetuses with a central nervous system defect is approximately 7-11%, according to studies(22,23). This is one of the most common congenital malformations associated with a pathogenic CNV. These genomic imbalances are most common in fetuses with Dandy-Walker malformation, cerebellar hypoplasia, or holoprosencephaly.
ACPA in fetuses with nucal hyperclarity
It was shown from the early 1990s that nucal hyperclarity is associated with a risk of chromosomal abnormalities such as trisomies 21, 18 and 13, triploidia, and gonosomal aneuploidia. It should be noted that trisomy 21 represents approximately 50% of the chromosomal abnormalities observed in the first quarter of nucal hyperclarity(24,25). More recently, several studies have shown the contribution of ACPA in this indication(16,23,26). In a recent meta-analysis of Grande et al. including 1,696 fetuses from 17 studies, the pathogenic CNV rate is 4% for isolated nucal hyperclarity and increases to 7% if another sign of ultrasound call is associated(27).
ACPA in fetuses with intrauterine growth retardation
It is known that most chromosomal abnormalities are associated with intrauterine growth retardation (IUCN). These include trisomy 18, trisomy 13, 4p16.3 (Wolf-Hirschhorn Syndrome), 5p15.2 (Cat Cree Syndrome) and 15q26 (including IGF1R gene)(28). With respect to the contribution of ACPA in this indication, the Shaffer et al. study showed that a pathogenic CNV was found in 2.7% of cases(23). This rate increases to about 10% if congenital malformation is associated with the NCIU. The recent meta-analysis of Borrell et al. revealed similar rates with about 4% pathogenic CNVs in isolated NCIU and about 10% if associated with congenital malformation(29). It is therefore entirely lawful to propose an ACPA in the case of CIDR. below the 3th percentile without identified etiology, even if no other fetal morphological abnormality is associated.
What type of ACPA should be used in prenatal?
The main obstacle to the application of ACPA in prenatal is the presence of SUVs and CNVs predisposing to pathologies that are difficult to predict phenotype (especially in the absence of ultrasound call signs, or in the case of minor ultrasound call signs). As a result, a reflection was made on the type of DNA chip to be used in antenatal: the objective was to detect the maximum number of pathogenic CNVs while limiting the detection of CNVs that raise genetic counseling problems. However, it has been shown that the resolution level of the chip used is correlated with the probability of detecting SUVs(30,31). As a result, some centers have chosen to adopt a different chip and/or with a different resolution than the one used in postnatal. In France, an enriched genomic chip on regions associated with known microdeletional or microduplicational syndromes and genes involved in severe diseases is often used(32). In addition, the detection threshold for a CNV was set at 1.5 Mb by most teams in France. In the future, the development of public databases (DECIPHER http://decipher.sanger.ac.uk/; Database of Genomic Variants http://projects.tcag.ca/variation/) or private will help clarify the clinical impact of some of these CNVs.
Conclusion
The advent of DNA chips is a major advance in the field of cytogenetics. Indeed, this tool offers the possibility of performing a penomic examination at a very high level of resolution. However, the detection of genomic imbalance sometimes raises many questions about its clinical consequences, which is a major limitation on the application of ACPA in prenatal diagnosis. However, ACPA is currently the preferred technique for fetal genome analysis when an invasive gesture for chromosome study is indicated.
*A reference genome is used for the CGH array but not for SNP chips.
References
Click on the references and access the Abstracts on

1. Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998 Oct 1;20(2):207–11. Search the abstract2. Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Döhner H, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997 Dec;20(4):399–407. Search the abstract3. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet [In Search the abstract4. Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet [Internet]. 2009 Feb;84(2):148–61. Search the abstract5. Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, et al. Gene Copy-Number Variation and Associated Polymorphisms of Complement Component C4 in Human Systemic Lupus Erythematosus (SLE): Low Copy Number Is a Risk Factor for and High Copy Number I Search the abstract6. Nakajima T, Kaur G, Mehra N, Kimura A. HIV-1/AIDS susceptibility and copy number variation in a gene encoding a natural ligand for HIV-1 co-receptor CCR5. Cytogenet Genome Res. 2009 Mar 11;123(1–4):156–60. Search the abstract7. Koolen DA, Vissers LELM, Pfundt R, de Leeuw N, Knight SJ, Regan R, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38(9):999–1001. Search the abstract8. Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008 Feb 14;358(7):667–75. Search the abstract9. Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, et al. Recurrent Rearrangements of Chromosome 1q21.1 and Variable Pediatric Phenotypes. N Engl J Med. 2008 Oct 16];359(16):1685–99. Search the abstract10. Green Snyder L, D’Angelo D, Chen Q, Bernier R, Goin-Kochel RP, Wallace AS, et al. Autism Spectrum Disorder, Developmental and Psychiatric Features in 16p11.2 Duplication. J Autism Dev Disord. 2016 Aug 21;46(8):2734–48. Search the abstract 11. Moreno-De-Luca A, Evans DW, Boomer KB, Hanson E, Bernier R, Goin-Kochel RP, et al. The Role of Parental Cognitive, Behavioral, and Motor Profiles in Clinical Variability in Individuals With Chromosome 16p11.2 Deletions. JAMA Psychiatry [Internet]. 20 Search the abstract12. Pichert G, Mohammed SN, Ahn JW, Ogilvie CM, Izatt L. Unexpected findings in cancer predisposition genes detected by array comparative genomic hybridisation: what are the issues? J Med Genet. 2011 Aug 1;48(8):535–9. Search the abstract13. South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet Med. 2013;15(11):901–9. Search the abstract14. Brady PD, Delle Chiaie B, Christenhusz G, Dierickx K, Van Den Bogaert K, Menten B, et al. A prospective study of the clinical utility of prenatal chromosomal microarray analysis in fetuses with ultrasound abnormalities and an exploration of a framewor Search the abstract15. Callaway JLA, Shaffer LG, Chitty LS, Rosenfeld JA, Crolla JA. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: A review of the literature. Prenat Diagn. 2013;33(12):11 Search the abstract16. de wit MC, Srebniak MI, Govaerts LCP, Van Opstal D, Galjaard RJH, Go ATJI. Additional value of prenatal genomic array testing in fetuses with isolated structural ultrasound abnormalities and a normal karyotype: a systematic review of the literature. U Search the abstract17. Song MS, Hu A, Dyhamenahali U, Chitayat D, Winsor EJT, Ryan G, et al. Extracardiac lesions and chromosomal abnormalities associated with major fetal heart defects: comparison of intrauterine, postnatal and postmortem diagnoses. Ultrasound Obstet Gyne Search the abstract18. Jansen FAR, Blumenfeld YJ, Fisher A, Cobben JM, Odibo AO, Borrell A, et al. Array comparative genomic hybridization and fetal congenital heart defects: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015 Jan;45(1):27–35. Search the abstract19. Donnelly JC, Platt LD, Rebarber A, Zachary J, Grobman WA, Wapner RJ. Association of Copy Number Variants With Specific Ultrasonographically Detected Fetal Anomalies. Obstet Gynecol. 2014 Jul;124(1):83–90. Search the abstract20. Yan Y, Wu Q, Zhang L, Wang X, Dan S, Deng D, et al. Detection of submicroscopic chromosomal aberrations by array-based comparative genomic hybridization in fetuses with congenital heart disease. Ultrasound Obstet Gynecol. 2014 Apr;43(4):404–12. Search the abstract21. ONKAR D, Onkar P, Mitra K. Evaluation of Fetal Central Nervous System Anomalies by Ultrasound and Its Anatomical Co-relation. J Clin DIAGNOSTIC Res. 2014 Jun;8(6):AC05-7. Search the abstract22. Sun L, Wu Q, Jiang S-W, Yan Y, Wang X, Zhang J, et al. Prenatal Diagnosis of Central Nervous System Anomalies by High-Resolution Chromosomal Microarray Analysis. Biomed Res Int. 2015:1–9. Search the abstract23. Shaffer LG, Rosenfeld JA, Dabell MP, Coppinger J, Bandholz AM, Ellison JW, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn. 2012;32(10):986–95. Search the abstract24. Nicolaides KH, Snijders RJ, Gosden CM, Berry C, Campbell S. Ultrasonographically detectable markers of fetal chromosomal abnormalities. Lancet (London, England). 1992 Sep 19;340(8821):704–7. Search the abstract25. Ville Y, Lalondrelle C, Doumerc S, Daffos F, Frydman R, Oury JF, et al. First-trimester diagnosis of nuchal anomalies: significance and fetal outcome. Ultrasound Obstet Gynecol. 1992 Sep 1;2(5):314–6. Search the abstract26. Leung TY, Vogel I, Lau TK, Chong W, Hyett JA, Petersen OB, et al. Identification of submicroscopic chromosomal aberrations in fetuses with increased nuchal translucency and apparently normal karyotype. Ultrasound Obstet Gynecol. 2011 Sep;38(3):314–9. Search the abstract27. Grande M, Jansen FAR, Blumenfeld YJ, Fisher A, Odibo AO, Haak MC, et al. Genomic microarray in fetuses with increased nuchal translucency and normal karyotype: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015 Dec;46(6):650–8. Search the abstract28. Salomon LJ, Malan V. Bilan étiologique du retard de croissance intra-utérin (RCIU). J Gynécologie Obs Biol la Reprod. 2013 Dec;42(8):929–40. Search the abstract29. Borrell A, Grande M, Pauta M, Rodriguez-Revenga L, Figueras F. Chromosomal Microarray Analysis in Fetuses with Growth Restriction and Normal Karyotype: A Systematic Review and Meta-Analysis. Fetal Diagn Ther. 2017;1–9. Search the abstract30. Oneda B, Baldinger R, Reissmann R, Reshetnikova I, Krejci P, Masood R, et al. High-resolution chromosomal microarrays in prenatal diagnosis significantly increase diagnostic power. Prenat Diagn. 2014;34(6):525–33. Search the abstract31. Cavalli P, Cavallari U, Novelli A. Array CGH in routine prenatal diagnosis practice. Prenat Diagn. 2012;32(7):708–9. Search the abstract32. Malan V, Lapierre J-M, Egloff M, Goidin D, Beaujard M-P, Maurin M-L, et al. A French Approach to Test Fetuses with Ultrasound Abnormalities Using a Customized Microarray as First-Tier Genetic Test. Cytogenet Genome Res. 2016 Jan 7;147(2–3):103–10. Search the abstract